初めに
lightGBMのハイパーパラメータのチューニングの仕方についてメモです。Optunaを使ってハイパーパラメータのチューニングを行います。
チューニングの方法を2種類試して、チューニングしない方法と比べてみます。最初にコード説明して、その後for loopで複数回実行してACCを比較してみます。
方法1 optunaのstudyを使用
方法2 optuna.integration.lightgbmを使う
方法3 チューニングなし
データ
心臓病のデータセット。心臓病かどうかを各種データから予測する。
https://www.kaggle.com/cherngs/heart-disease-cleveland-uci
参考:https://archive.ics.uci.edu/ml/datasets/Heart+Disease
Authors:
1. Hungarian Institute of Cardiology. Budapest: Andras Janosi, M.D.
2. University Hospital, Zurich, Switzerland: William Steinbrunn, M.D.
3. University Hospital, Basel, Switzerland: Matthias Pfisterer, M.D.
4. V.A. Medical Center, Long Beach and Cleveland Clinic Foundation:Robert Detrano, M.D., Ph.D.
前処理 データを加工
from sklearn.model_selection import train_test_split as tts
import lightgbm as lgb
import pandas as pd
df = pd.read_csv("heart_cleveland_upload.csv")
train_validation,test = tts(df,test_size=0.2,random_state=1)
train,validation = tts(train_validation,test_size=0.2)
# target variableが最後の列にあるので、xから除きyに入れる
x_train_validation = train_validation.iloc[:,:-1].to_numpy()
y_train_validation = train_validation.iloc[:,-1:].to_numpy().ravel()
x_train = train.iloc[:,:-1].to_numpy()
y_train = train.iloc[:,-1:].to_numpy().ravel()
x_val = validation.iloc[:,:-1].to_numpy()
y_val = validation.iloc[:,-1:].to_numpy().ravel()
x_test = test.iloc[:,:-1].to_numpy()
y_test = test.iloc[:,-1:].to_numpy().ravel()
# lightGBM用のデータに変換する
lgb_train_validation = lgb.Dataset(x_train_validation,y_train_validation, free_raw_data=False)
lgb_train = lgb.Dataset(x_train, y_train, free_raw_data=False)
lgb_eval = lgb.Dataset(x_val, y_val, free_raw_data=False)lgb用のデータ変換する際に、free_raw_data=Falseを入れています。これがないと、次のステップで以下のようなエラーがでます。
LightGBMError: Reducingmin_data_in_leafwithfeature_pre_filter=truemay cause unexpected behaviour for features that were pre-filtered by the largermin_data_in_leaf.
You need to setfeature_pre_filter=falseto dynamically change themin_data_in_leaf.
[LightGBM] [Fatal] Reducingmin_data_in_leafwithfeature_pre_filter=truemay cause unexpected behaviour for features that were pre-filtered by the largermin_data_in_leaf.
You need to setfeature_pre_filter=falseto dynamically change themin_data_in_leaf.
free_raw_data=Falseの意味は、If you set free_raw_data=True (default), the raw data (with Python data struct) will be freed.と書かれています。正直よくわかりませんが、エラーを回避するにはfalseです。
方法1 optunaのstudyを使用
import optuna
import numpy as np
import sklearn
import lightgbm as lgb
# studyの際に、最大化する数値をreturnする関数 ※1
def objective(trial):
param = {
'objective': 'binary',
'metric': trial.suggest_categorical('metric', ['binary_error',"binary_logloss"]),
'lambda_l1': trial.suggest_loguniform('lambda_l1', 1e-8, 10.0),
'lambda_l2': trial.suggest_loguniform('lambda_l2', 1e-8, 10.0),
'num_leaves': trial.suggest_int('num_leaves', 2, 256),
'feature_fraction': trial.suggest_uniform('feature_fraction', 0.4, 1.0),
'bagging_fraction': trial.suggest_uniform('bagging_fraction', 0.4, 1.0),
'bagging_freq': trial.suggest_int('bagging_freq', 1, 7),
'min_child_samples': trial.suggest_int('min_child_samples', 5, 100),
}
gbm = lgb.train(param, lgb_train,valid_sets=lgb_eval)
preds = gbm.predict(x_val)
pred_labels = np.rint(preds)
accuracy = sklearn.metrics.accuracy_score(y_val, pred_labels)
return accuracy
# optunaのstudyを実施。accuracyの方向をmaximizeする。
study = optuna.create_study(direction='maximize')
study.optimize(objective, n_trials=1000)
# 上のstudyで得られた最高のパラメータを取り出す
bestparams = study.best_trial.params
# 上のstudyで得られた最高のパラメータには、
# 単一選択肢だったものは保存されないので、
# 消えてしまったobjectiveを加える
bestparams["objective"]="binary"
# 改めて最高のパラメータでtrain_validation dataを学習
model = lgb.train(bestparams,lgb_train_validation)
pred = model.predict(x_test)
pred = (pred > 0.5) * 1
print("method1",bestparams)
print("ACC", sklearn.metrics.accuracy_score(pred,y_test))method1 {‘metric’: ‘binary_logloss’, ‘lambda_l1’: 0.4258483883300017, ‘lambda_l2’: 0.03231662719797015, ‘num_leaves’: 213, ‘feature_fraction’: 0.707809231580766, ‘bagging_fraction’: 0.8061561588920214, ‘bagging_freq’: 2, ‘min_child_samples’: 26, ‘objective’: ‘binary’}
ACC 0.8666666666666667
※1 設定できるパラメータ・optunaの関数について
設定できるパラメータはlightGBMの公式サイトを参照
https://lightgbm.readthedocs.io/en/latest/Parameters.html
‘objective’: regression, regression_l1, huber, fair, poisson, quantile, mape, gamma, tweedie, binary, multiclass, multiclassova, cross_entropy, cross_entropy_lambda, lambdarank, rank_xendcg
‘metric’: mae, mse, rmse, mape, binary_logloss,binary_error, multi_logloss, multi_error, cross_entropy, cross_entropy_lambda
optunaの関数については、optunaのサイトを参照
https://optuna.readthedocs.io/en/stable/reference/generated/optuna.trial.Trial.html
以下の関数を使うことで上記のパラメータを動かしてくれます。
suggest_categorical(name, choices)・・・Suggest a value for the categorical parameter.
suggest_discrete_uniform(name, low, high, q)・・・Suggest a value for the discrete parameter.
suggest_float(name, low, high, *[, step, log])・・・Suggest a value for the floating point parameter.
suggest_int(name, low, high[, step, log])・・・Suggest a value for the integer parameter.
suggest_loguniform(name, low, high)・・・Suggest a value for the continuous parameter.
suggest_uniform(name, low, high)・・・Suggest a value for the continuous parameter.
方法2 optuna.integration.lightgbmを使う
import optuna.integration.lightgbm as LGB_optuna
param = {
'objective': 'binary',
'metric': 'binary_error',
}
# なぜか1回学習させたら、もう一度データを初期化しないといけない※
lgb_train = lgb.Dataset(x_train, y_train,free_raw_data=False)
lgb_eval = lgb.Dataset(x_val, y_val,free_raw_data=False)
# パラメータ選ばせる
best = LGB_optuna.train(param, lgb_train,valid_sets=lgb_eval)
# 選んだパラメータで学習
model = lgb.train(best.params, lgb_train_validation)
pred = model.predict(x_test)
pred = (pred > 0.5) * 1
print("method2",best.params )
print( "ACC",sklearn.metrics.accuracy_score(pred,y_test))
method2 {‘objective’: ‘binary’, ‘metric’: ‘binary_error’, ‘feature_pre_filter’: False, ‘lambda_l1’: 8.558465348052447, ‘lambda_l2’: 3.424010078618614e-08, ‘num_leaves’: 31, ‘feature_fraction’: 0.52, ‘bagging_fraction’: 0.4920018288227101, ‘bagging_freq’: 3, ‘min_child_samples’: 20, ‘num_iterations’: 1000, ‘early_stopping_round’: None}
ACC 0.85
ACCは方法1(0.86)より若干低下しました。
※ なぜかデータを初期化しないと、以下のようなエラーがでてクラッシュします。方法1で一回データを使ってしまって変化が起きたのでしょうか。
[LightGBM] [Fatal] Reducingmin_data_in_leafwithfeature_pre_filter=truemay cause unexpected behaviour for features that were pre‑filtered by the largermin_data_in_leaf.
You need to setfeature_pre_filter=falseto dynamically change themin_data_in_leaf.
[LightGBM] [Fatal] Cannot change feature_pre_filter after constructed Dataset handle.
方法3 チューニングなし
param = {
'objective': 'binary',
'metric': 'binary_error',
}
model = lgb.train(param, lgb_train,valid_sets=lgb_eval)
pred = model.predict(x_test)
pred = (pred > 0.5) * 1
print("method3",model.params)
print("ACC", sklearn.metrics.accuracy_score(pred,y_test))method3 {‘objective’: ‘binary’, ‘metric’: ‘binary_error’, ‘num_iterations’: 100, ‘early_stopping_round’: None}
ACC 0.7666666666666667
ACCは下がりました。
ちなみに、今までのパラメータをまとめると以下のようになります。デフォルトの設定も調べてみました。
method1 {
‘objective’: ‘binary’,
‘metric’: ‘binary_logloss’,
‘lambda_l1’: 0.4258483883300017,
‘lambda_l2’: 0.03231662719797015,
‘num_leaves’: 213,
‘feature_fraction’: 0.707809231580766,
‘bagging_fraction’: 0.8061561588920214,
‘bagging_freq’: 2,
‘min_child_samples’: 26
}
method2 {
‘objective’: ‘binary’,
‘metric’: ‘binary_error’,
‘feature_pre_filter’: False,
‘lambda_l1’: 8.558465348052447,
‘lambda_l2’: 3.424010078618614e-08,
‘num_leaves’: 31,
‘feature_fraction’: 0.52,
‘bagging_fraction’: 0.4920018288227101,
‘bagging_freq’: 3,
‘min_child_samples’: 20,
‘num_iterations’: 1000,
‘early_stopping_round’: None
}
method3 {
‘objective’: ‘binary’,
‘metric’: ‘binary_error’,
‘num_iterations’: 100,
‘early_stopping_round’: None
}
defalt{
‘objective’: ‘binary’,
‘metric’: ‘binary_error’,
‘feature_pre_filter’: true,
‘lambda_l1’: 0.0,
‘lambda_l2’: 0.0,
‘num_leaves’: 31,
‘feature_fraction’: 1.0,
‘bagging_fraction’: 1.0,
‘bagging_freq’: 0,
‘min_child_samples’: 20,
‘num_iterations’: 100,
‘early_stopping_round’: 0
}
10回実施 どの方法がいいか
それでは、1回の結果だけだと信頼性がないのでtrain_test_splitのseedを変化させて10回実施します。そして、どの方法が一番良かったかを計算します。
from sklearn.model_selection import train_test_split as tts
import lightgbm as lgb
import pandas as pd
import optuna
import numpy as np
import sklearn
import optuna.integration.lightgbm as LGB_optuna
acc_lightgbm_m1 = []
acc_lightgbm_m2 = []
acc_lightgbm_m3 = []
for i in range(10):
############## creation of data for lightgbm ##################
df = pd.read_csv("heart_cleveland_upload.csv")
train_validation,test = tts(df,test_size=0.2,random_state=i)
train,validation = tts(train_validation,test_size=0.2)
x_train_validation = train_validation.iloc[:,:-1].to_numpy()
y_train_validation = train_validation.iloc[:,-1:].to_numpy().ravel()
x_train = train.iloc[:,:-1].to_numpy()
y_train = train.iloc[:,-1:].to_numpy().ravel()
x_val = validation.iloc[:,:-1].to_numpy()
y_val = validation.iloc[:,-1:].to_numpy().ravel()
x_test = test.iloc[:,:-1].to_numpy()
y_test = test.iloc[:,-1:].to_numpy().ravel()
lgb_train_validation = lgb.Dataset(x_train_validation,y_train_validation,free_raw_data=False)
lgb_train = lgb.Dataset(x_train, y_train,free_raw_data=False)
lgb_eval = lgb.Dataset(x_val, y_val,free_raw_data=False)
############ method 1 ##############
def objective(trial):
param = {
'objective': 'binary',
'metric': trial.suggest_categorical('metric', ['binary_error',"binary_logloss"]),
'lambda_l1': trial.suggest_loguniform('lambda_l1', 1e-8, 10.0),
'lambda_l2': trial.suggest_loguniform('lambda_l2', 1e-8, 10.0),
'num_leaves': trial.suggest_int('num_leaves', 2, 256),
'feature_fraction': trial.suggest_uniform('feature_fraction', 0.4, 1.0),
'bagging_fraction': trial.suggest_uniform('bagging_fraction', 0.4, 1.0),
'bagging_freq': trial.suggest_int('bagging_freq', 1, 7),
'min_child_samples': trial.suggest_int('min_child_samples', 5, 100),
}
gbm = lgb.train(param, lgb_train,valid_sets=lgb_eval)
preds = gbm.predict(x_val)
pred_labels = np.rint(preds)
accuracy = sklearn.metrics.accuracy_score(y_val, pred_labels)
return accuracy
study = optuna.create_study(direction='maximize')
study.optimize(objective, n_trials=1000)
bestparams = study.best_trial.params
bestparams["objective"]="binary"
model = lgb.train(bestparams, lgb_train_validation)
pred = model.predict(x_test)
pred = (pred > 0.5) * 1
acc_lightgbm_m1.append(sklearn.metrics.accuracy_score(pred,y_test))
############ method 1 ##############
############ method 2 ##############
param = {
'objective': 'binary',
'metric': 'binary_error',
}
lgb_train = lgb.Dataset(x_train, y_train,free_raw_data=False)
lgb_eval = lgb.Dataset(x_val, y_val,free_raw_data=False)
best = LGB_optuna.train(param, lgb_train,valid_sets=lgb_eval)
model = lgb.train(best.params, lgb_train_validation)
pred = model.predict(x_test)
pred = (pred > 0.5) * 1
acc_lightgbm_m2.append(sklearn.metrics.accuracy_score(pred,y_test))
################ method 2 ######################
################ method 3 ######################
param = {
'objective': 'binary',
'metric': 'binary_error',
}
model = lgb.train(param, lgb_train,valid_sets=lgb_eval)
pred = model.predict(x_test)
pred = (pred > 0.5) * 1
acc_lightgbm_m3.append(sklearn.metrics.accuracy_score(pred,y_test))
import matplotlib.pyplot as plt
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
ax.boxplot([acc_lightgbm_m1, acc_lightgbm_m2, acc_lightgbm_m3],
labels=['Method1', 'Method2', 'Method3'])
ax.set_ylabel('Accuracy')
ax.set_ylim(0, 20)
plt.style.use('ggplot')
plt.show()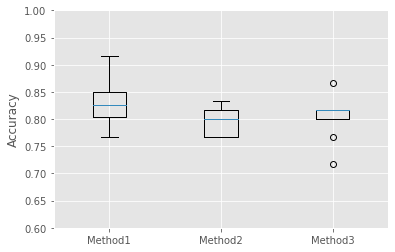
Method1の方がよさそうですね。統計的には??
ここからはRで。。。
ANOVAで3群間の差を調べます。
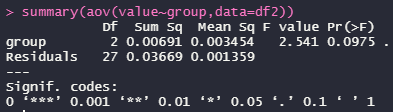
3群間で差なし。もちろんpairwise t testでも差なし。
だめなんだろうけど単純に、method1とmethod2の平均の差をtテスト、method1とmethod3のtテストをやりたい。
method1とmethod2 p-value = 0.04367
method1とmethod3 p-value = 0.1356
あんまりハイパーパラメータチューニング意味ない??
お知らせ
新しい職場を探しています。私を雇用したいと思った方は、お問い合わせフォームからご連絡をお願い致します。
